Cluster
SSH connection
Once you have an account on the platform, to connect to our cluster or other command line tools, you need to connect
to our bastion server genossh.genouest.org.
This is the entrypoint to our network. From there, you can submit jobs with the Slurm job manager or connect to other resources.
You first need to connect to the front-end server via SSH from your computer.
You can connect to genossh.genouest.org from anywhere, but only with a properly configured SSH Key.
ed25519 keys cannot be used for connecting to GenOuest, please make sure to use an RSA key.
Genouest-generated SSH keys
A key-generating tool is available in the GenOuest account manager, in the SSH tab.
By clicking on the "Generate new key" button, a new keypair will be created. The public key will be automatically added on our server. Make sure to download the private rsa key (for linux or linux subsystem) or putty private key (for windows). You can also save the public key if you wish to use the keypair elsewhere.
Please note that the generated key will not use a passphrase.
From Windows
Putty
On Windows, Putty can be used to load SSH keys and connect via SSH to the cluster. Have a look at this video tutorial explaining the whole procedure (creating an SSH key and then connecting to the cluster). (If you used the Genouest tool, you can skip the parts about key generation and adding the public key to the interface.)
MobaXterm
MobaXterm is another available tool to connect to the cluster.
If you did not use the Genouest tool, you can use MobaXterm to generate your SSH keys :
- Head to
Tools -> MobaKeyGen - Create a 4096 RSA key (with an optional passphrase)
- Save the public and private key files
- Copy the public key from the interface or the public key file
- Paste it to https://my.genouest.org (
SSH -> Public SSH key)
To connect to our server, you will need to add your private key (either the GenOuest-generated one, or your own) to MobaXterm itself :
- Head to
Settings -> Configuration -> SSH tab - Tick
Use internal SSH agent "MobAgent" - Use the + button to select your private key
- Restart MobaXterm
You should then be able to connect using ssh <your-login>@genossh.genouest.org.
Linux subsystem
If you prefer, you can also use the Linux subsystem of Windows (in this case, see the Linux/Mac paragraph below).
From Linux or Mac
Creating your key
If you did not create a keypair using the GenOuest tool, you will first need to generate an SSH key. To do so, use this command on your computer:
ssh-keygen -t rsa -b 4096
The command will ask for a password: it will protect your SSH key, and you will need it everytime you will use it to connect to the cluster (depending on your configuration, a program named ssh-agent can remember this password after you entered it the first time you connect).
The ssh-keygen program will create two files in your home directory:
$HOME/.ssh/id_rsa
$HOME/.ssh/id_rsa.pub
id_rsa is your private key: keep this file secret.
id_rsa.pub is your public key. You need to open this file and copy-paste its contents to https://my.genouest.org (SSH -> Public SSH key form on the right side, once your are logged in).
Connecting to Genouest
Add your key in your ssh agent (this is not always needed depending on your configuration):
ssh-add $HOME/.ssh/id_rsa
You should then be able to connect to the cluster with this command:
ssh <your-login>@genossh.genouest.org
Once connected, to access other servers, you might still need to use your ssh key.
- either copy your private key on the cluster:
scp $HOME/.ssh/id_rsa <your-login>@genossh.genouest.org:~/.ssh/ - or allow agent forwarding when connecting to the cluster:
ssh -A <your-login>@genossh.genouest.org
Data transfers
Protocols
It is possible to copy data from/to GenOuest using multiple protocols.
Warning:
Access to /scratch has been temporarily removed from FTP pending resolution of a bug with our scratch storage system. You can use genossh with sftp/ssh to transfer your data to/from scratch.
| server | protocols | storages |
|---|---|---|
| genossh.genouest.org | scp, rsync, sftp | /home, /projects, /scratch |
| openstack.genouest.org | scp, rsync, sftp | /home, /projects, /scratch |
| ftp.genouest.org | ftp, sftp | /home, /projects, /scratch |
| data-access.cesgo.org | https | /home |
| homeandco.genouest.org | https | /home, /projects |
FTP
example of FTP configuration with FileZilla :
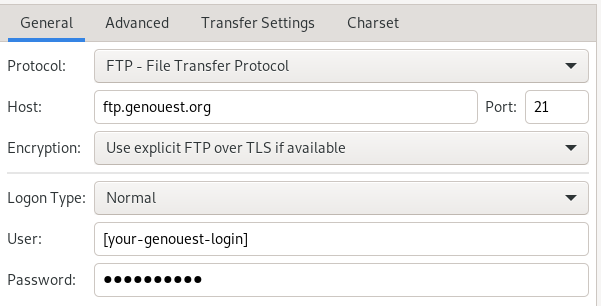
You can use any ftp compliant tool to transfer data, be sure however to use the ftp protocol (secure, TLS in explicit mode)
SFTP
/!\ You will need to use an ssh key for SFTP.
example of SFTP configuration with FileZilla :
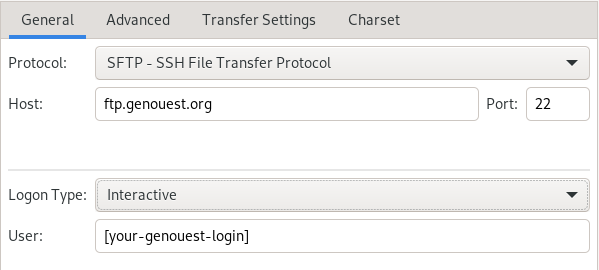
Interactive login type is used with ssh agent. You can select the Key file instead.
Data storage
You have access to three volumes, available on all computing nodes.
Home directory
Your home directory (/home/genouest/your-group/your-login). We have a total of around 100TB of storage capacity shared between all the home directories, and each user has a quota of 120GB. You can check your disk usage with this command:
quota -s
A snapshot mechanism is available on this volume, in case you erased a file by mistake. See below.
Giving file access to someone else
On UNIX systems, it is possible to make a file or directory publicly available server-wide. Since June 2021, the default policy for this type of shares is more restrictive : nobody except the owners can access their home directory and its content.
To make an item available you should :
- First: allow users to step into your home
chmod o+x /home/genouest/<your-group>/<your-login>
- Then: give access to your item following the appropriate method
For a file :
chmod o+r /home/genouest/<your-group>/<your-login>/<your-file>
For a directory :
find /home/genouest/<your-group>/<your-login>/<your-directory> -type d | xargs chmod o+rx
find /home/genouest/<your-group>/<your-login>/<your-directory> -type f | xargs chmod o+r
Now, any people logged onto the cluster can access this item.
Project directory
A project directory (/projects/your-group) that you share with your team. We have a total of around 200TB of storage capacity shared between all these group directories.
Each project has a specific quota, and a single person in your team is responsible to grant you access to this volume.
You can check your disk usage with the command:
df -h /projects/<your-group>
A snapshot mechanism is available on this volume, in case you erased a file by mistake. See below.
To join an existing group, please contact the group owner, owners can manage their projects to add or remove members.
To ask for a new project directory, connect to our account manager in my project and request a new project.
Important:
All data in a project directory is owned by the project owner and remains under his sole responsability. Project owners may copy/move/delete any data in the project directory, whoever the file creator was.
Access to data by a non project member must be approved by the project owner.
Scratch directory
A high performance storage space (/scratch/your-login). Each user has a quota of 250GB. You can check your disk usage with the command:
df -h /scratch/<your-login>
Good practices
Quotas are intentionally restrictive, if you need them to be increased, please contact support@genouest.org.
As a general rule, users should not write in the /home or /projects directories during a job, nor do any heavy read operations on these volumes. They are used to keep your data safe. During jobs, one should use the /scratch directory. This directory is hosted by a high performance system and is designed for temporary data. It supports heavy read and write operations.
Please note that none of your data is backed up. If you would like us to backup your data for specific reasons, you can contact us and we will help you to find a solution.
We strongly advise you to anticipate your storage needs: if you plan to generate a large amount of data, please contact us before to check that we have the possibility to host this data. It is preferable to anticipate this when applying for grants that imply data generation and analysis.
Before generating data on the cluster, please do not forget to check the remaining available space
Snapshots
If you erase some files by mistake, you can recover the files by looking in a special .snapshot directory, available both in any /home or /projects sub-directory.
The snapshots are performed each hour and are kept for 5 weeks. To access the snapshot files of your account just go to the .snapshot directory.
cd .snapshot
There, you will see several directories in which copies of your files at different times are stored.
The directories are easily recognizable by their name: hourly, daily, weekly. Snapshots for the 'home' directory include hourly (last 24 hours), daily (last 5 days, excluding weekends), and weekly (last 5 weeks). Snapshots for the 'groups' directory include daily and weekly, kept for the same duration as the 'home' directory. There are no snapshots for the 'scratch' directory.
Please note that snapshots are not backups. They provide protection against user error, but not against physical failure of the data storage servers.
Please consider an external backup solution if your data is valuable.
Software
Preinstalled catalog
Pre-installed software are available in /softs/local (see software manager for a list of installed software). To use a software, you have to load its environment. For example to load python 2.7 you can launch this command (the dot at the beginning is important):
. /softs/local/env/envpython-2.7.sh
This will automatically configure your shell environment to execute the selected tool. Any subsequent python command you will launch will use this 2.7 version.
To get a list of all environments available, just list the content of /softs/local/env/env*, or look at the list on software manager.
Note: DO NOT USE the python/perl/... of the node directly, always load a specific version from /softs/local.
Conda
Conda is a system allowing package, dependency and environment management for any programming language. It is widely used to install software on the cluster.
Conda is a way to create custom environments, completely isolated from the other software installed on the cluster. You can install all the Conda packages you want in each isolated Conda environment.
A list of available conda packages is here: https://anaconda.org/anaconda/repo.
Conda allows you to install the software you need in your own storage volumes (/home, /projects or /scratch). The software needs to be available as Conda packages.
By default, the channels bioconda and conda-forge are enabled
on the cluster. The Bioconda channel in particular is tailored for
bioinformatics tools. You may add other channels if you need. Please keep in mind that
private channels might present security risks (software will not be vetted).
If possible, please keep to the standard channels.
https://docs.conda.io/projects/conda/en/latest/user-guide/tasks/manage-channels.html
Due to licensing restrictions from the Anaconda company, we no longer enable the default channel on our installation. It shouldn't be a problem for most users though as the bioconda and conda-forge channels are usually enough for most bioinformatics tools.
To use Conda, first source it the usual way (on a compute node):
. /local/env/envconda.sh
With Conda, you can create as many environments as you want, each one containing a list of packages you need. You need to activate an environment to have access to software installed in it.
To create a new environment containing biopython, deeptools (v2.3.4),
bowtie and blast, run the following command (replace ~/my_env by a path where you want to store the environment):
conda create -p ~/my_env biopython deeptools=2.3.4 bowtie blast
To activate it:
conda activate ~/my_env
To deactivate it:
conda deactivate
Although it's not recommended, you can activate multiple environments by adding the --stack option. In this case, the last activated environment will have a higher priority than the other ones.
conda activate --stack ~/my_env
We have also installed Mamba as an alternative to Conda. It is a reimplementation written in C++ designed to be much faster. To use it, just source the conda env as usual (. /local/env/envconda.sh), then you can replace the conda commands by mamba such as in this example:
mamba create -p ~/my_env biopython deeptools=2.3.4 bowtie blast
As of 2024-01-04, we have installed Conda version 23.11 that now uses Mamba's algorithm internally. This means that the conda command should be just as fast as mamba now. Although you should now use conda in most cases, we've left mamba installed for compatibility.
Activation and deactivation of environments still needs to be done with the conda activate command.
Quota exceeded with Conda
Conda stores some cached data inside a hidden directory in your HOME directory (~/.conda/). If you install a lot of different packages, the size of this directory will grow, and might hit your quota limit on your home.
To free up space by deleting unused cached data, you can run the following command, which will show you what data will be deleted, and ask for confirmation:
conda clean --all
Virtualenv
Several versions of Python are available on the cluster. Each one comes with a specific set of modules preinstalled. If you need to install a module, or to have a different module version, you can use Virtualenv. Virtualenvs are a way to create a custom Python environment, completely isolated from the Python installation on the cluster. You can install all the Python modules you want in this isolated environment.
To use it, first create a new virtualenv like this:
. /local/env/envpython-3.7.6.sh
virtualenv ~/my_new_env
This will create the directory ~/my_new_env. This directory will contain a minimal copy of Python 3.7.6 (the one you sourced just before), without any module installed in it, and completely isolated from the global 3.7.6 python version installed by GenOuest. If you prefer to use a Python 2.7 version, you can source 2.7.15 of Python if you prefer:
. /local/env/envpython-2.7.15.sh
virtualenv ~/my_new_env
To use this virtualenv, you need to activate it:
. ~/my_new_env/bin/activate
Once activated, your prompt will show that you activated a virtualenv:
(my_new_env)[login@cl1n025 ~]$
You can then install all the Python modules you need in this virtualenv:
pip install biopython
pip install pyaml...
Now, when you run python, you will be using the virtualenv’s Python version, containing only the modules you installed in it.
Once you have finished working with the virtualenv, you can stop using it and switch back to the normal environment like this:
deactivate
You can create as many virtualenv as you want, each one being a directory that you can safely remove when you don’t need it anymore.
Reference databanks
GenOuest maintains several up-to-date databanks, available in /db from every computation node. The complete list of maintained databanks is available here
Please refer to the BioMAJ page for more information regarding databanks.
Submitting jobs with Slurm
See the Slurm section